Genetically engineered control systems
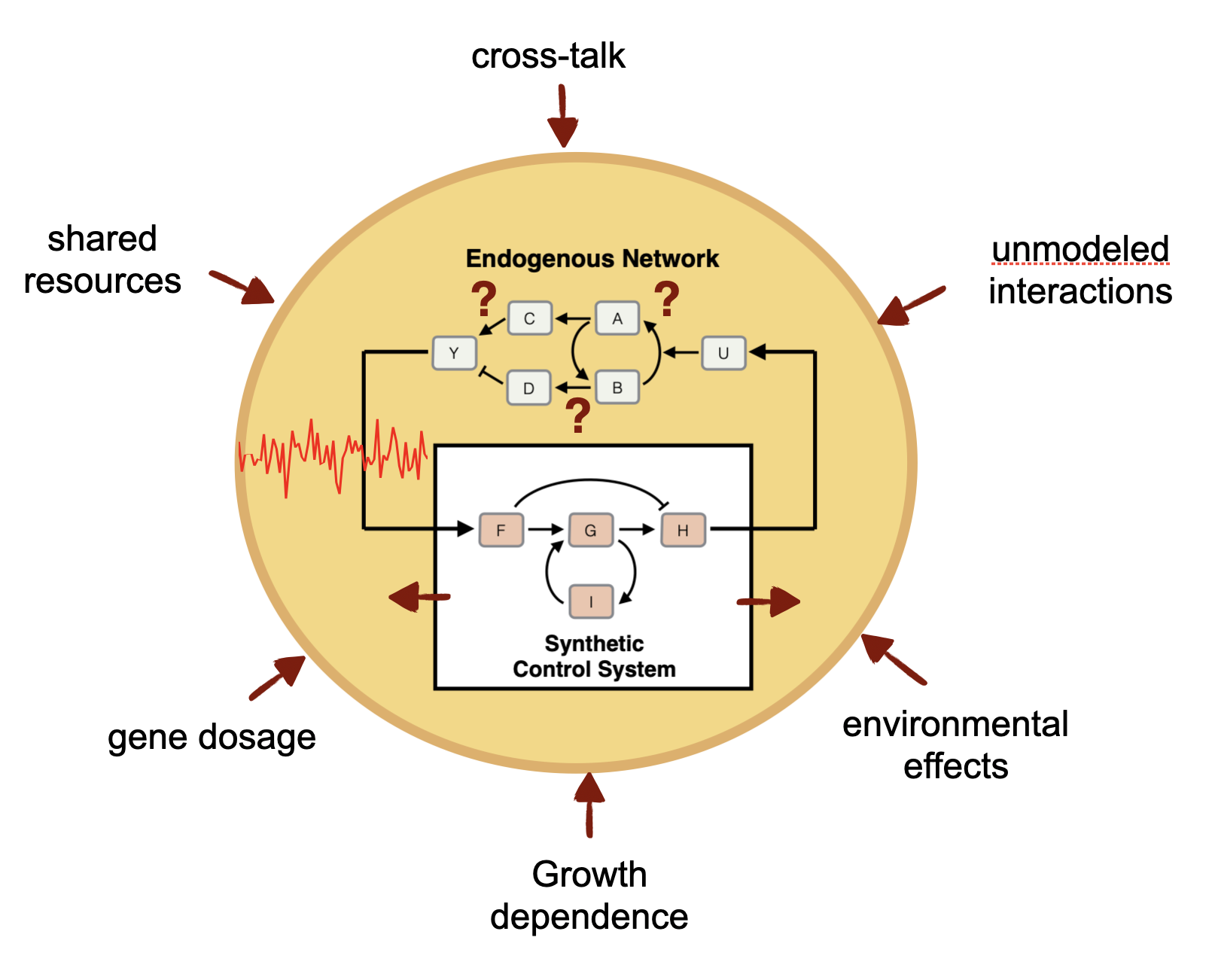
A genetic control system must contend with many challenges, including uncertainty in the toplogy and parameters of the biomolecular process to be controlled, persistent environmental disturbances, and strong dependence on in the highly variable cellular context in which the controller functions. On top of that, the molecular controller must function reliably in the extremely noisy environment of the cell including the noise generated from its own molecular reactions. The resulting genetic feedback control systems implemented are autonomous, highly dynamic, and generally stochastic in nature. We are developing the foundations of a control theory needed to analyze and design such biomolecular controllers. In the lab, we are constructing these controllers in E. coli, yeast, and mammalian cells.
A Universal Integral Control System for Robust Perfect Adaptation
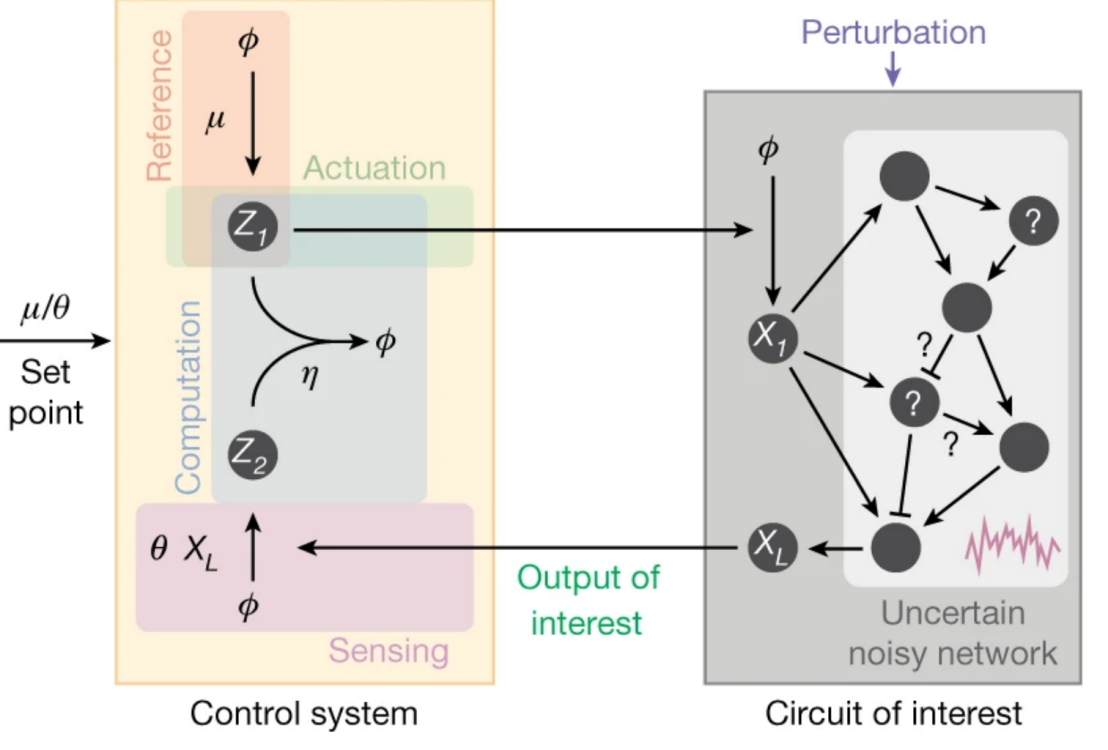
We proved mathematically that there is a single fundamental biomolecular controller topology that realizes integral feedback and achieves robust perfect adaptation in arbitrary intracellular networks with noisy dynamics. This toplogy requires the interaction of two antagnistic molecules closing the feedback loop. We call this strategy antithetic integral control.
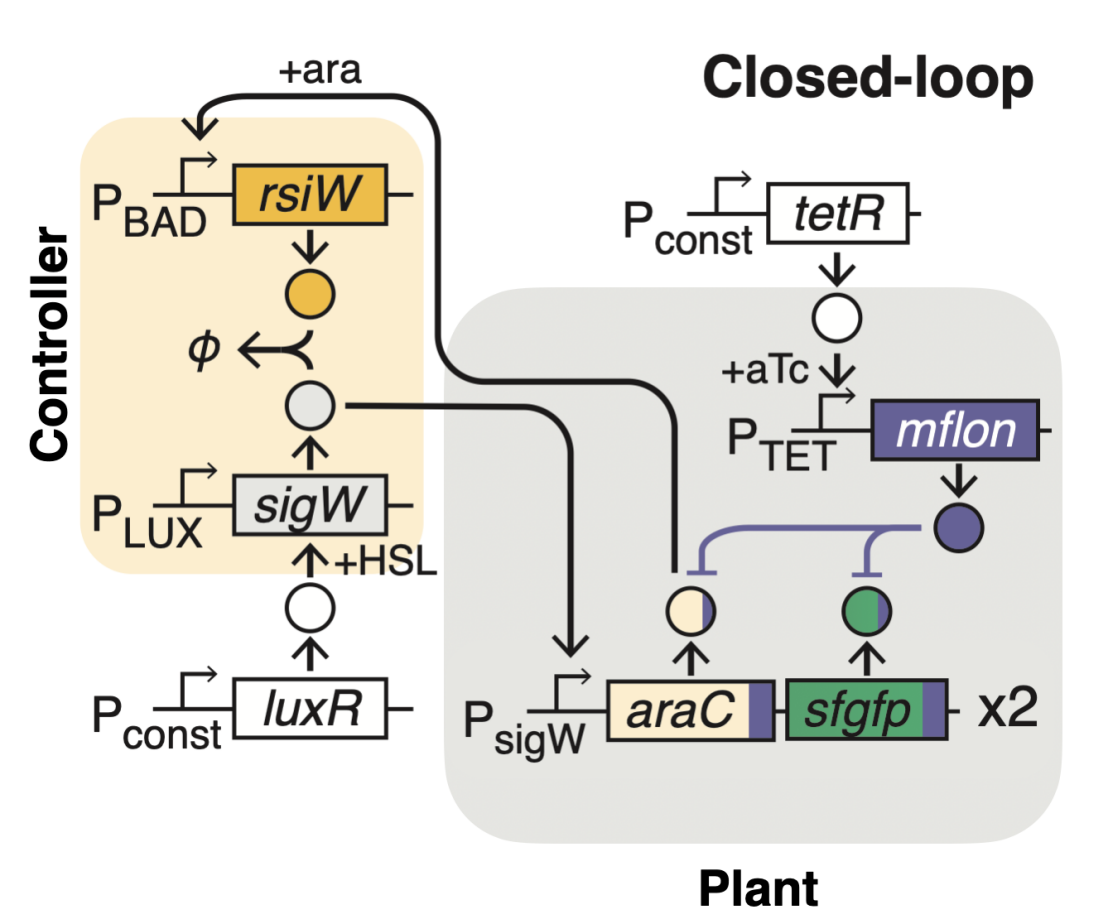
On the basis of this concept, we genetically engineered the first synthetic integral feedback controller in living cells and demonstrated its tunability and adaptation properties. A growth-rate control application in E. coli shows the intrinsic capacity of our integral controller to deliver robustness and highlights its potential use as a versatile controller for regulation of biological variables in uncertain networks.
High Performance Genetic Controllers for Mammalian Cells
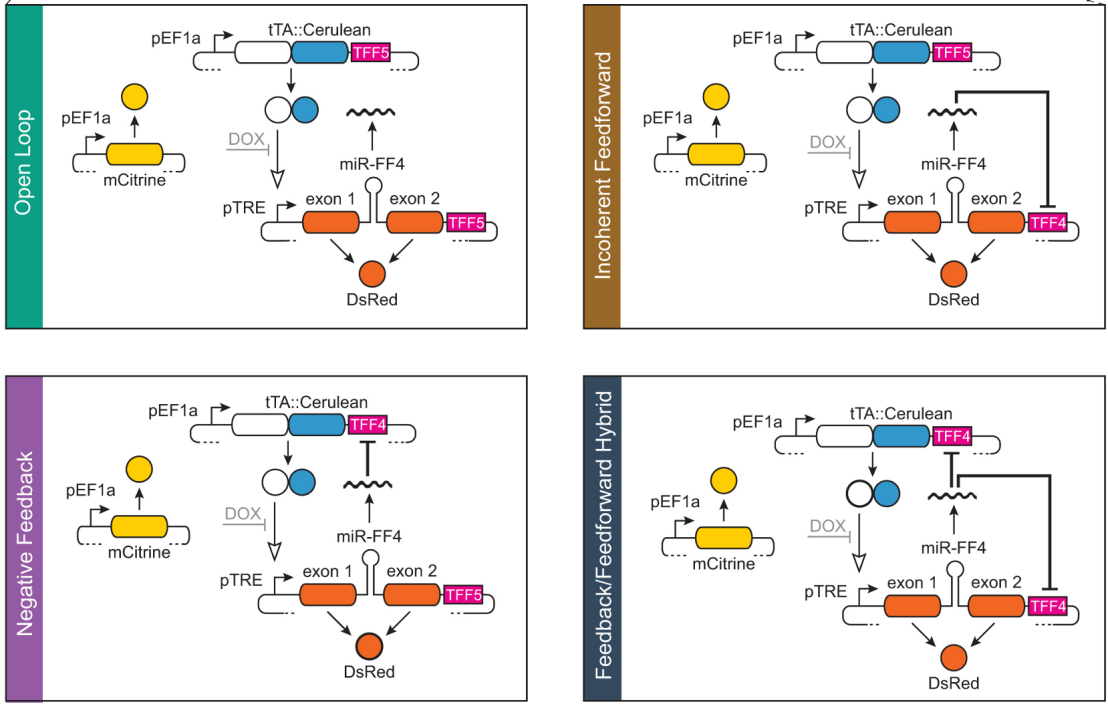
Tunable induction of gene expression is an essential tool in biology and biotechnology. We introduced a novel family of miRNA gene expression control systems of varying complexity with significantly enhanced performance. We demonstrated the benefits of these controllers in two applications in a culture of CHO cells for protein manufacturing and in human-induced pluripotent stem cells.
Molecular Circuits for Dynamic Noise Filtering
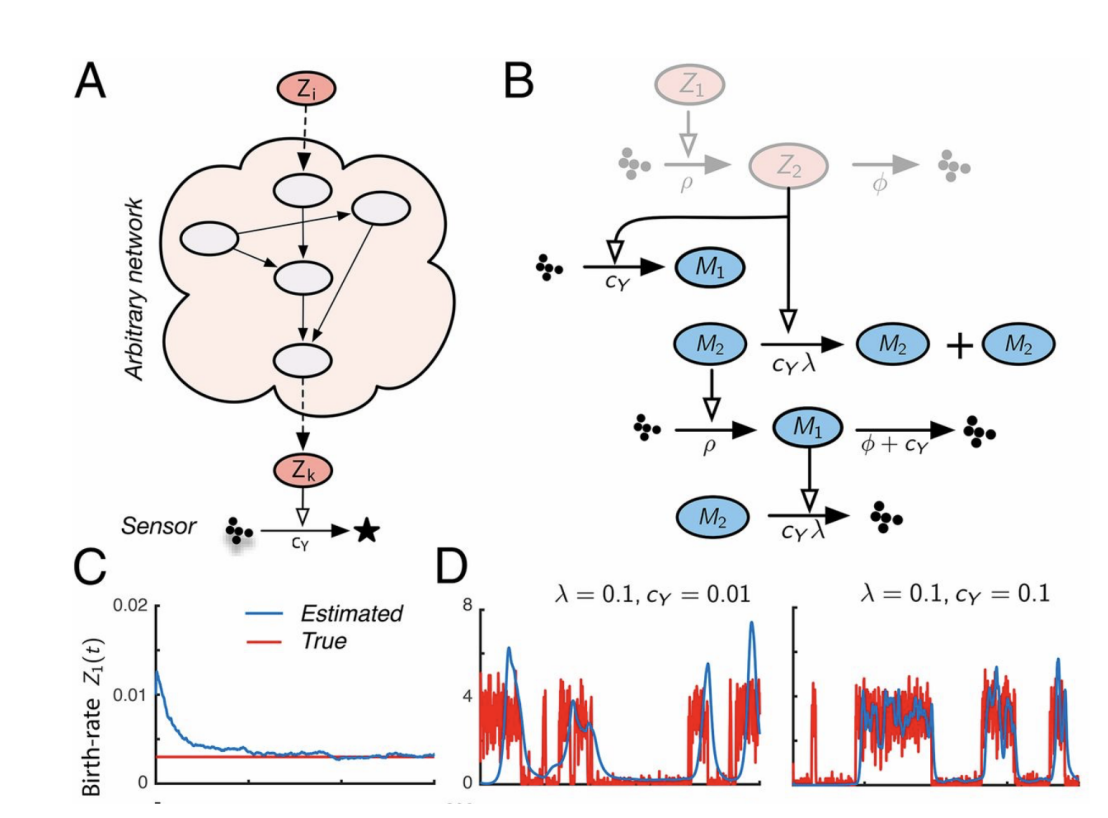
The invention of the Kalman filter is a crowning achievement of filtering theory that has revolutionized technology. A similar protocol to deal with noise in synthetic biology has been missing. We developed an optimal filtering theory for noisy biochemical networks and showed how the resulting filters can be implemented at the molecular level. We experimentally demonstrated our findings in vitro and in vivo.
Integral Control for Optimal Productivity in a Biofuel Metabolic Pathway
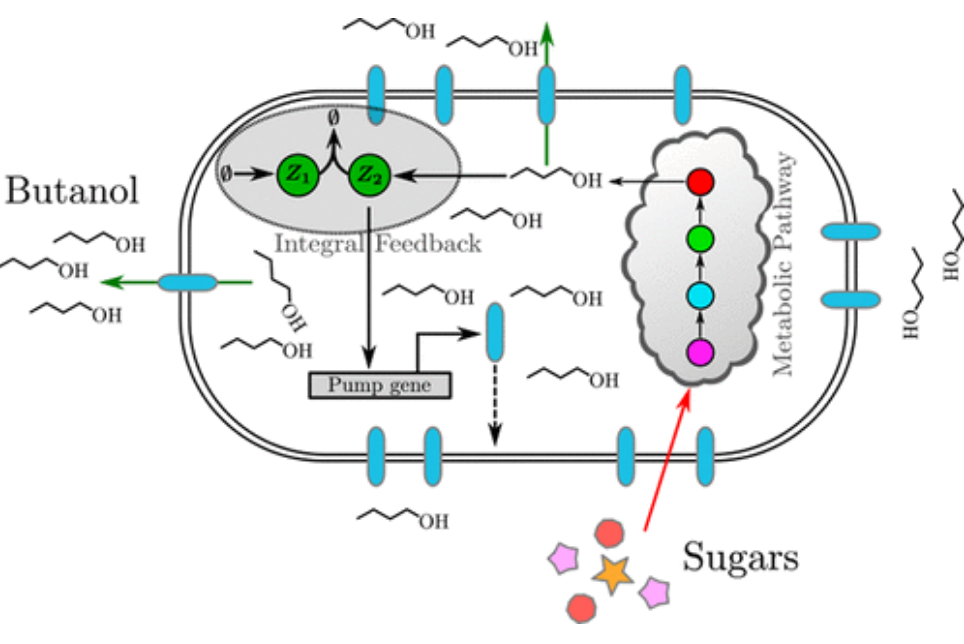
The production of complex biomolecules by genetically engineered organisms is one of the most promising applications of metabolic engineering and synthetic biology. We showed that an antithetic integral control strategy achieves robust perfect adaptation and a high robust rate of production of extracellular biofuel, even when the parameters of the model and controller are poorly known and implemented.
Stephanie K. Aoki, Gabriele Lillacci, Ankit Gupta, Armin Baumschlager, David Schweingruber & Mustafa Khammash. "A universal biomolecular integral feedback controller for robust perfect adaptation," Nature. 19 June 2019. external pagedoi.org/10.1038/s41586-019-1321-1call_made
Gabriele Lillacci, Yaakov Benenson and Mustafa Khammash. "Synthetic control systems for high performance gene expression in mammalian cells," Nucleic Acids Research, vol. 46: no. 18, pp. 9855-9863, Oxford: Oxford University Press (OUP), 2018.external pageDOI: 10.1093/nar/gky795call_made
Corentin Briat, Ankit Gupta and Mustafa Khammash. "Antithetic proportional-integral feedback for reduced variance and improved control performance of stochastic reaction networks," Journal of the Royal Society. Interface, 15 (143): 20180079, London: The Royal Society, 2018. external pagedoi:10.1098/rsif.2018.0079call_made
Corentin Briat and Mustafa Khammash. "Perfect Adaptation and Optimal Equilibrium Productivity in a Simple Microbial Biofuel Metabolic Pathway Using Dynamic Integral Control," ACS Synthetic Biology, 7 (2): 419-431, Washington, DC: American Chemical Society, 2018. external pagedoi:10.1021/acssynbio.7b00188call_made
C. Briat, C. Zechner, and M. Khammash. "Design of a Synthetic Integral Feedback Circuit: Dynamic Analysis and DNA Implementation," ACS Synthetic Bioliogy, 2016. external pagedoi:10.1021/acssynbio.6b00014call_made
C. Zechner, G. Seelig, M. Rullan, and M. Khammash. "Molecular circuits for dynamic noise filtering," Proceedings of the National Academy of Sciences of the United States of America, 2016. external pagedoi:10.1073/pnas.1517109113call_made
C. Briat, A. Gupta, and M. Khammash. "Antithetic Integral Feedback Ensures Robust Perfect Adaptation in Noisy Bimolecular Networks," Cell Systems, 2016. external pagedoi:http://dx.doi.org/10.1016/j.cels.2016.01.004call_made
Andreas Milias-Argeitis, Stefan Engblom, Pavol Bauer and Mustafa Khammash. "Stochastic focusing coupled with negative feedback enables robust regulation in biochemical reaction networks," Interface: Journal of the Royal Society, vol. 12: no. 113, pp. 20150831, London: Royal Society, 2015. external pageDOI: 10.1098/rsif.2015.0831call_made
A. Gupta, C. Briat, and M. Khammash, "A Scalable Computational Framework for Establishing Long-Term Behavior of Stochastic Reaction Networks," PLoS Computational Biology, 2014. external pagedoi:http://dx.doi.org/10.1371/journal.pcbi.1003669call_made
