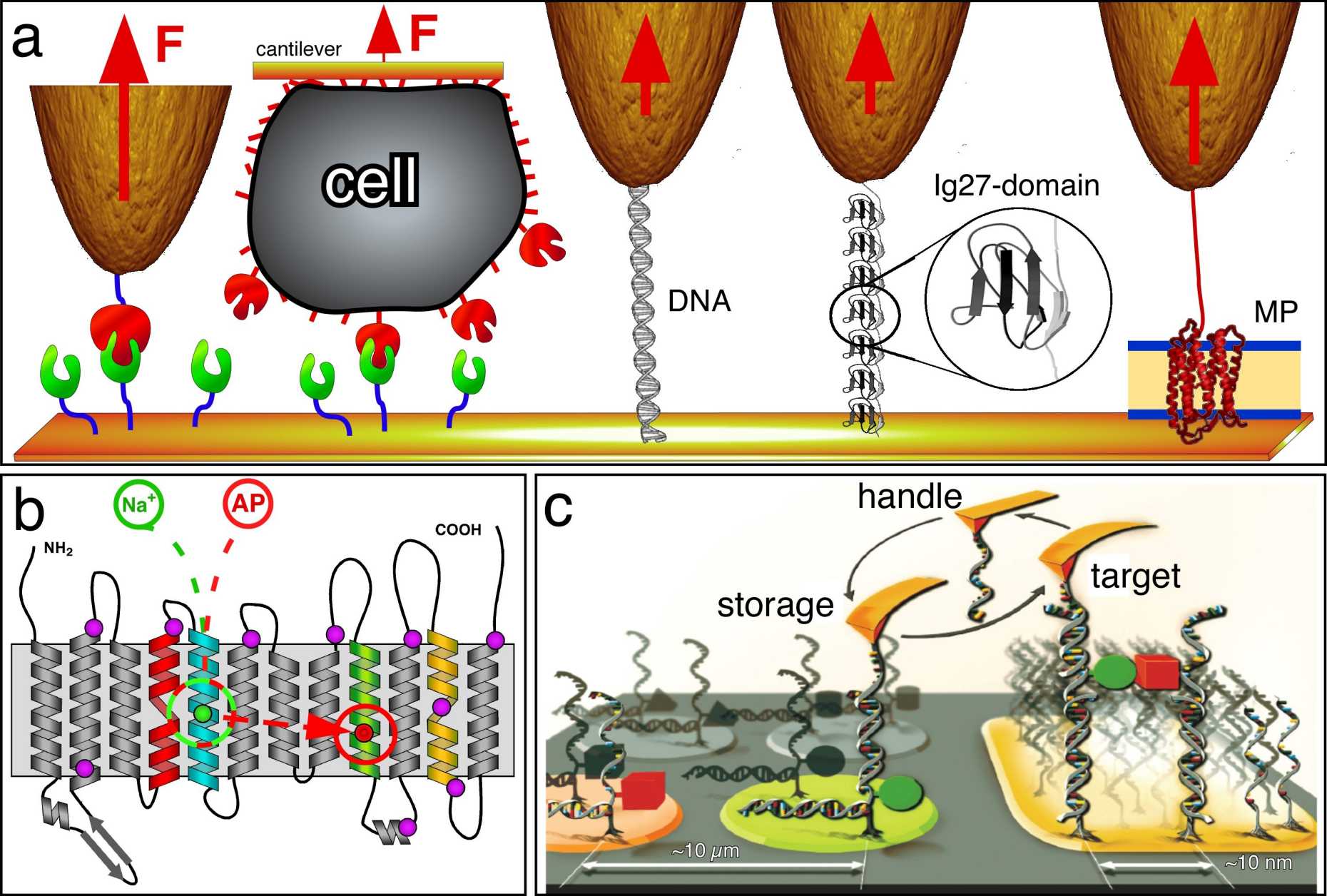
Molecular Force Spectroscopy
Molecular interactions drive all processes in life. They determine the molecular crosstalk and build the basic language of biological processes. Molecular interactions fold the polypeptide into the functional protein, stabilize the structure, or lead to protein misfolding. These molecular forces determine protein-protein interactions, switching on and off ion channels, ligand-binding, the functional states of receptors, and the supramolecular assembly of molecular machines to functional units. On more complex scales molecular interactions guide cell adhesion, migration, communication and differentation. Other examples show how they sculpture reaction pathways of biological processes. Because of this enormous importance it is one pertinent demand in life sciences to characterize how these interactions are established and how they guide biological processes ranging from the molecular to cellular scale. To decipher fundamentals of this biological language at the molecular scale, we have pioneered atomic force microscopy (AFM)-based single-molecule force spectroscopy (SMFS) of native membrane proteins.
By applying SMFS to membrane proteins we characterize their unfolding and folding pathways into the lipid or native membrane at physiological relevant conditions (e.g., buffer solution, room temperature or 37°C). Alternatively we also apply SMFS to quantify and structurally localize inter- and intramolecular interactions of membrane proteins. The structural localization allows to map these interactions to single alpha-helices, beta-strands or polypeptide loops.
Further reading
YidC assists the stepwise and stochastic folding of membrane proteins
T. Serdiuk, D. Balasubramaniam, J. Sugihara, S.A. Mari, H.R. Kaback & D.J. Muller
Nature Chemical Biology (2016) 11, 911-917.
Molecular plasticity of the human voltage-dependent anion channel embedded into a membrane
L. Ge, S. Villinger, S.A. Mari, K. Giller, C. Griesinger, S. Becker, D.J. Müller & M. Zweckstetter
Structure (2016) 24, 585-594.
Quantifying and identifying two ligand-binding sites while imaging native human membrane receptors by AFM
M. Pfreundschuh, D. Alsteens, R. Wieneke, C. Zhang, S.R. Coughlin, R. Tampé, B.K. Kobilka & D.J. Müller
Nature Communications (2015) 6, 8857. external pageonlinecall_made
Impact of holdase chaperones Skp and SurA on the folding of β-barrel outer membrane proteins
J. Thoma, B.M. Burmann, S. Hiller & D.J. Muller
Nature Structural and Molecular Biology (2015), 22, 795-802.
Single-molecule force spectroscopy of membrane proteins from membranes freely spanning across nanoscopic pores
R. Petrosyan, C.A. Bippes, S. Walheim, D. Harder, D. Fotiadis, T. Schimmel, D. Alsteens & D.J. Müller
Nano Letters (2015) 15, 3624-3433.
Dynamic single-molecule force spectroscopy of rhodopsin in native membranes
P.S. Park & D.J. Muller
Methods in Molecular Biology (2015) 1271, 173-185.
Observing a lipid-dependent alteration in single lactose permeases
T. Serdiuk, J. Sugihara, S.A. Mari, H.R. Kaback & D.J. Müller
Structure (2015) 23, 754-761.
Action of the Hsp70 chaperone system observed with single proteins
J.M. Nunes, M. Mayer-Hartl, F.U. Hartl & D.J. Muller
Nature Communications (2015) 6, 6307.
Substrate-induced changes in the structural properties of LacY
T. Serdiuk, M.G. Madej, J. Sugihara, S. Kawamura, S.A. Mari, H.R. Kaback & D.J. Muller
Proc. Natl. Acad. Sci. USA (2014) 111, E1571-E1580.
The peptide transporter DtpA populates two alternate conformations from which inhibitor binding promotes one
C. Bippes, L. Ge, M. Meury, D. Harder, Z. Ucurum, H. Daniel, D. Fotiadis & D.J. Muller
Proc. Natl. Acad. Sci. USA (2013) 110, E3978-3986.
Mechanistic explanation of different unfolding behaviors observed for transmembrane and soluble β-barrel proteins
U. Hensen & D.J. Muller
Structure (2013) 21, 1317-1324.
Single-molecule force spectroscopy of G-protein coupled receptors
M. Zocher, Ch. Bippes, C. Zhang & D.J. Muller
Chemical Society Reviews (2013) 42, 7801-7815.
Nanomechanical properties of proteins and membranes depend on loading rate and electrostatic interactions
I. Medalsy & D.J. Muller
ACS NANO (2013) 7, 2642-2650.
Structural, kinetic, and mechanical differences between dark-state rhodopsin and opsin
S. Kawamura, M. Gerstung, L. Colozo, J. Helenius, N. Beerenwinkel, P.S.H. Park & D.J. Muller
Structure (2013) 21, 426-437.
Cholesterol increases kinetic, energetic, and mechanical stability of the human β2 adrenergic receptor
M. Zocher, C. Zhang, G.F.S. Rassmussen, B.K. Kobilka & D.J. Muller
Proc. Natl. Acad. Sci. USA (2012) 109, E3463-3473.
Out but not in: β-strands shape the unfolding pathway but not the refolding of the large transmembrane β-barrel protein FhuA
J. Thoma, P. Bosshart, M. Pfreundschuh & D.J. Muller
Structure (2012) 20, 2185-2190.
Characterization of co-translationally formed nanodisc complexes with small multidrug transporters, proteorhodopsin and with the E. coli MraY translocase
C. Roos, M. Zocher, D.J. Muller, D. Munch, T. Schneider, H.G. Sahl, F. Scholz, J. Wachtveitl, Y. Ma, D. Proverbio, E. Henrich, V. Dötsch & F. Bernhard
Biochemical Biophysical Acta (2012) 1818, 3098-3106.
Ligand-specific interactions modulate kinetic, energetic and mechanical properties of the human β2 adrenergic receptor
M. Zocher, J.J. Fung, B.K. Kobilka & D.J. Muller
Structure (2012) 20, 1391-1402.
Structural, energetic and mechanical perturbations in a rhodopsin mutant that causes congenital stationary night blindness
S. Kawamura, A.L. Colozo, L. Ge, D.J. Muller & P.S.H. Park
Journal of Biological Chemistry (2012) 287, 21826-21835.
Single-molecule force spectroscopy from nanodiscs – an assay to quantify folding, stability and interactions of native membrane proteins
M. Zocher, C. Roos, S. Wegmann, P. Bosshart, V. Doetsch, F. Bernhard & D.J. Muller
ACS NANO (2012) 6, 961-971.
KpOmpA a transmembrane protein anchoring the outer membrane of Klebsiella pneumoniae unfolds and refolds in response to tensile load
P. Bosshart, I. Iordanov, C. Garzon-Coral, P. Demange, A. Engel, A. Milon & D.J. Muller
Structure (2012) 20, 121-127.
Locating an extracellular K+-dependent interaction site that modulates betaine-binding of the Na+-coupled betaine symporter BetP
L. Ge, C. Perez, I. Waclavska, C. Ziegler & D.J. Muller
Proc. Natl. Acad. Sci. USA (2011) 108, E890-898.
Competing interactions stabilize pro- and anti-aggregant conformations of human Tau
S. Wegmann, J. Schöler, C.A. Bippes, E. Mandelkov & D.J. Muller
Journal of Biological Chemistry (2011) 286, 20512-20524.
One β-hairpin after the other: Exploring refolding pathways and kinetics of the transmembrane β-barrel protein OmpG
M. Damaghi, S. Koester, C.A. Bippes, Ö. Yildiz & D.J. Muller
Angewandte Chemie International Edition (2011) 50, 7422-7424.
High-resolution atomic force microscopy and spectroscopy of native membrane proteins
Ch. Bippes & D.J. Muller
Reports on Progress in Physics (2011) 74, 086601.
Five challenges to bringing single-molecule force spectroscopy into the living cell
Y.F. Dufrene, E. Evans, A. Engel, J. Helenius, H.E. Gaub & D.J. Müller
Nature Methods (2011) 8, 123-127.
Conservation of molecular interactions stabilizing bovine and mouse rhodopsin
S. Kawamura, A.T. Colozo, D.J. Müller & P.S.H. Park
Biochemistry (2010) 49, 10412-10420.
A ”force buffer” protecting immunoglobulin titin
J.A. Nunes, U. Hensen, L. Ge, M. Lipinsky, J. Helenius, H. Grubmüller & D.J. Müller
Angewandte Chemie International Edition (2010) 49, 3528-3531.
Assessing the structure and function of single biomolecules with scanning transmission electron and atomic force microscopes
S. Muller, D.J. Müller & A. Engel
Micron (2010) 42, 186-195.
Dual energy landscape: The functional state of the outer-membrane β-barrel protein OmpG molds its unfolding energy landscape
M. Damaghi, K.T. Sapra, S. Köster, Ö. Yildiz, W. Kühlbrandt & D.J. Müller
Proteomics (2010) 10, 4151-4162.
Quantifying interactions that switch the β-barrel membrane protein OmpG from the open to the closed state
M. Damaghi, C.A. Bippes, S. Köster, S.A. Mari, Ö. Yildiz, W. Kühlbrandt & D.J. Müller
Journal of Molecular Biology (2010) 397, 878-882.
Probing the interactions of carboxy-atractyloside and atractyloside with the yeast mitochondrial ADP/ATP carrier
A. Kedrov, A.M. Hellawell, A. Klosin, R.B. Broadhurst, E.R.S. Kunji & D.J. Müller
Structure (2010) 18, 39-46.
