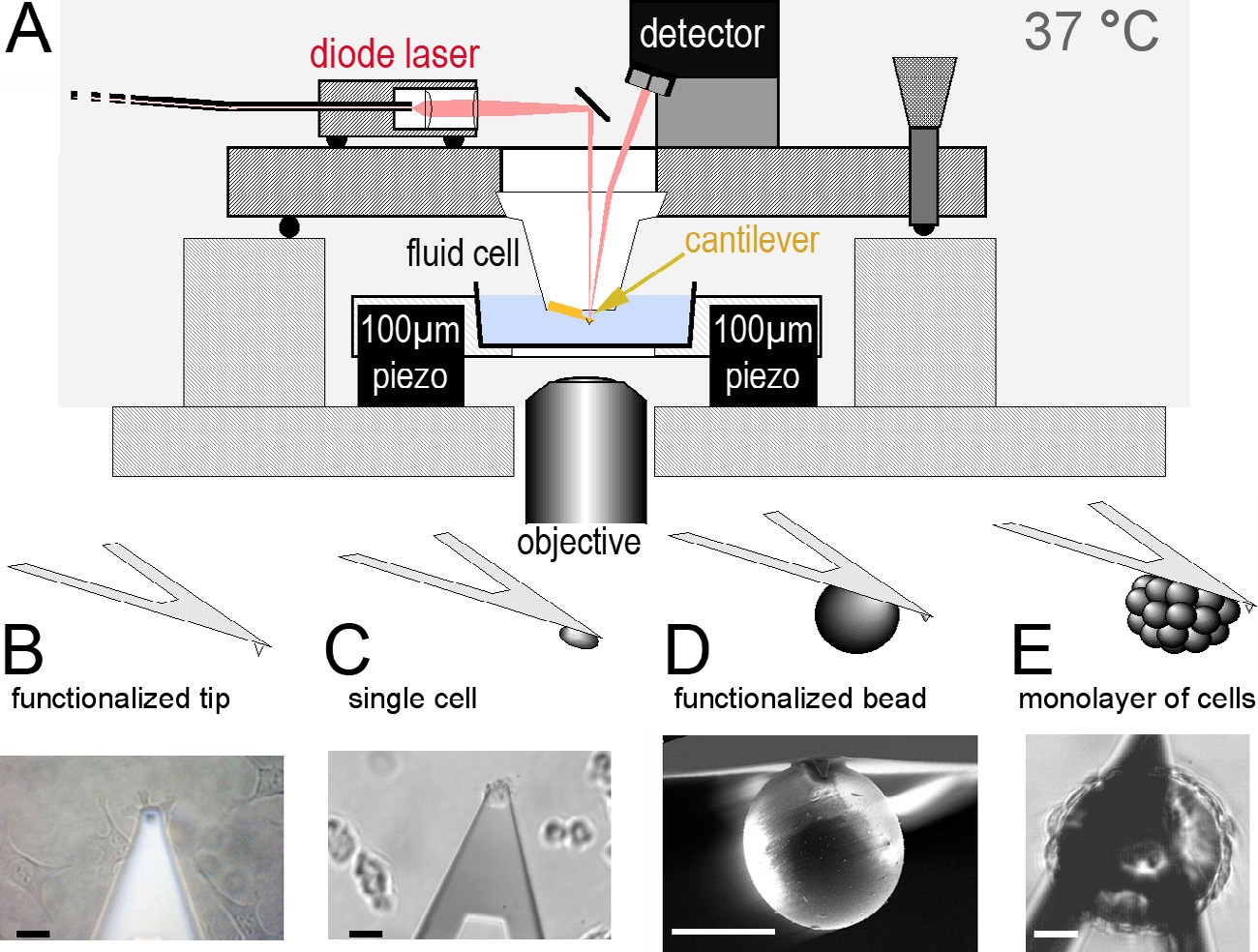
Cellular Force Spectroscopy
A broad spectrum of biological processes requires controlled cell adhesion, including embryonic development, assembly of tissues and the nervous system, cellular communication, inflammation and wound healing, tumor metastasis, cell culturing, and viral and bacterial infection. Although much is known about cell adhesion, many questions remain unanswered owing to its multiple facets and complexity. Cell adhesion is commonly defined as the binding of a cell to a substrate, which can be another cell, a surface or an organic matrix. The process is regulated by specific cell-adhesion molecules (CAMs), which are typically transmembrane receptors that comprise an intracellular domain that interacts with cytoplasmic proteins, including the cytoskeleton, and an extracellular domain that specifically binds to adhesion partners. However, the molecular mechanisms by which CAMs work and how they regulate different types of adhesion are open debates. For example, an extensive array of proteins is known to be involved in adhesive assemblies, i.e. focal adhesions [cell–extracellular-matrix (ECM) junctions], but the contributions of these proteins to the strength of adhesion are not quantitatively understood. To understand cell adhesion, therefore, the vast amount of qualitative data that is available must be augmented with quantitative data of the physics of adhesion. Therefore, to obtain quantiative insight into how cells initiate and strengthen adhesion, we develop AFM-based single-cell force spectroscopy (SCFS) tools and apply these tools to very mammalian systems.
Further reading
Kindlin-2 cooperates with talin to activate integrins and induces cell spreading by directly binding paxillin
M. Theodosiou, M. Widmaier, R.T. Böttcher, E. Rognoni, M. Veelders, M. Bharadwaj, A. Lambacher, K. Austen, D.J. Müller, R. Zent & R. Fässler
eLife (2016) 5, e10130. external pageonlinecall_made
In PC3 prostate cancer cells ephrin receptors crosstalk to β1-integrins to strengthen adhesion to collagen type I
M. Yu, J. Wang, D.J. Muller & J. Helenius
Scientific Reports (2015) 5, 8206.
Assay for characterizing the recovery of vertebrate cells for adhesion measurements by single-cell force spectroscopy
R. Schubert, N. Strohmeyer, M. Bharadwaj, R. Subramaniam, M. Krieg, J. Friedrichs, C.M. Franz & D.J. Müller
FEBS Letters (2014) 588, 3639-3648.
Dynamic coupling of ALCAM to the actin cortex strengthens cell adhesion to CD6
J. te Riet, J. Helenius, N. Strohmeyer, A. Cambi, C.G. Fidgor & D.J. Muller
Journal of Cell Science (2014) 127, 1595-1606.
Single-cell force spectroscopy, an emerging tool to quantify cell adhesion to biomaterials
A. Taubenberger, D. Hutmacher & D.J. Muller
Tissue Engineering (2014) 20, 40-55.
αv-integrins are required for mechanotransduction in MDCK epithelial cells
T.P. Teräväinen, S.M. Myllymäki, J. Friedrichs, N. Strohmeyer, J.V. Moyano, C. Wu, K.S. Matlin, D.J. Muller & A. Manninen
PLoS One (2013) 8, e71485.
Deciphering teneurin domains that facilitate cellular recognition, cell-cell adhesion and neurite outgrowth using AFM-based single-cell force spectroscopy
J. Beckmann, R. Schubert, R. Chiquet-Ehrismann & D.J. Muller
Nano Letters (2013) 13, 2937-2946.
A practical guide to quantify cell adhesion using single-cell force spectroscopy
J. Friedrichs, K.R. Legate, R. Schubert, M. Bharadwaj, C. Werner, D.J. Muller & M. Benoit
Methods (2013) 60, 169–178.
Quantifying cellular adhesion to covalently immobilized extracellular matrix proteins by single-cell force spectroscopy
J. Friedrichs, C. Werner & D.J. Muller
Methods in Molecular Biology (2013) 1046, 19-37.
Force nanoscopy of living cells
D.J. Müller & Y.F. Dufrene
Current Biology (2011) 21, R212-R216.
Planar cell polarity signalling coordinates epithelial organ formation by regulating cell adhesion properties in progenitor cells
P. Oteiza, M. Köppen, M. Krieg, S. Preibisch, E. Pulgar, C. Farias, C. Melo, D.J. Müller, M. Tada, S. Hartel, C.-P. Heisenberg & M.L. Concha
Development (2010) 137, 3459-68.
Control of directed cell migration in vivo by membrane-to-cortex attachment
A. Diz-Muñoz, M. Krieg, M. Bergert, I. Ibarlucea-Benitez, D.J. Müller, E. Paluch, & C.P. Heisenberg
PLoS Biology (2010) 8, e1000544.
Blood vessel lumen formation via electrostatic repulsion
B. Strilić, J. Eglinger, M. Krieg, M. Zeeb, J. Axnick, P. Babál, D.J. Müller & E. Lammert
Current Biology (2010) 20, 2003-2009.
Quantifying cellular adhesion to extracellular matrix components by single-cell force spectroscopy
J. Friedrichs, J. Helenius & D.J. Müller
Nature Protocols (2010) 5, 1353-1361.
Stimulated single-cell force spectroscopy to quantify cell adhesion receptor crosstalk
J. Friedrichs, J. Helenius & D.J. Müller
Proteomics (2010) 10, 1455-1462.
Effect of unlocking RGD-motifs in collagen I on pre-osteoblast adhesion and differentiation
A.V. Taubenberger, M.A. Woodruff, H. Bai, D.J. Müller & D. Hutmacher
Biomaterials (2010) 31, 2827-2835.
Alignment and cell-matrix interactions of human corneal endothelial cells on nanostructured collagen type I matrices
S. Ulbrich, J. Friedrichs, M. Valtink, S. Murovski, C.M Franz, D.J. Müller, R.H.W. Funk & K. Engelmann
Investigative Opthamology & Visual Science (2010) 51, 6303-6310.
Movement directionality in collective germ layer progenitor migration
Y. Arboleda-Estudillo, M. Krieg, J. Stuhmer, D.J. Müller & C.P. Heisenberg
Current Biology (2010) 20, 161-169.
Force probing surfaces of living cells to molecular resolution
D.J. Müller, J. Helenius, D. Alsteens & Y.F. Dufrene
Nature Chemical Biology (2009) 5, 383-390.
New frontiers in atomic force microscopy: Analyzing interactions from single-molecules to cells
D.J. Müller, M. Krieg, D. Alsteens & Y.F. Dufrene
Current Opinion in Biotechnology (2009) 20, 4-13.
TPA primes α2β1 integrins for cell adhesion
M. Tulla, J. Helenius, D.J. Müller & J. Heino
FEBS Letters (2008) 582, 3520-3524.
A bond for a lifetime: Employing membrane nanotubes from living cells to determine receptor-ligand kinetics
M. Krieg, J. Helenius, C.-P. Heisenberg & D.J. Müller
Angewandte Chemie International Edition (2008) 47, 9775 –9777.
Single-cell force spectroscopy
J. Helenius, C.P. Heisenberg, H.E. Gaub & D.J. Müller
Journal of Cell Science (2008) 121, 1785-1791.
Galectin-3 regulates the kinetics of integrin α2β1 mediated adhesion to collagen-I and -IV
J. Friedrichs, A. Manninen, D.J. Müller & J. Helenius
Journal of Biological Chemistry (2008) 283, 32264-32272.
Single-cell force spectroscopy reveals β1-integrin as a central molecule mediating adhesion of 32D-BCR-ABL cells to bone marrow stroma cells
F.A. Fierro, A. Taubenberger, P.-H. Puech, G. Ehninger, M. Bornhäuser, D.J. Müller & T. Illmer
Journal of Molecular Biology (2008) 377, 1082-1093.
Atomic force microscopy as a multifunctional molecular toolbox in nanobiotechnology
D.J. Müller & Y. Dufrene
Nature Nanotechnology (2008) 3, 261-269.
Quantifying adhesive and tensile cell properties determining germ layer organization during gastrulation
M. Krieg, Y. Arboleda, P.-H. Puech, J. Kaefer, F. Graener, D.J. Müller & C.P. Heisenberg
Nature Cell Biology (2008) 10, 429-436.
Studying integrin-mediated cell adhesion on the single-molecule level by using AFM force spectroscopy
C.M. Franz, A. Taubenberger, P.-H. Puech & D.J. Müller
Science STKE (2007) 406, pl5.
Galectin-3 and galectin-9 contribute to adhesion and cystogenesis in epithelial MDCK cells
J. Friedrichs, J. Torkko, J. Helenius, J. Füllekrug, D.J. Müller, K. Simons & A. Manninen
Journal of Biological Chemistry (2007) 282, 29375-29383.
Revealing early steps of α2β1 integrin-mediated adhesions to collagen type I using single-cell force spectroscopy
A. Taubenberger, D.A. Cisneros, J. Friedrichs, P.-H. Puech, D.J. Müller & C.M. Franz
Molecular Biology of the Cell (2007) 18, 1634-1644.
A new technical approach to quantify cell adhesion forces by AFM
P.-H.Puech, K. Poole, D. Knebel & D.J. Müller
Ultramicroscopy (2006) 106, 637-644.
Wnt11 functions in gastrulation by controlling cell cohesion through Rab5c and E-cadherin
F. Ulrich, M. Krieg, E. Schötz, V. Link, I. Castanton, A. Taubenberger, D.J. Müller, P.-H. Puech & C.P. Heisenberg
Developmental Cell (2005) 9, 555-564.
Analysing focal adhesion structure by AFM
C. Franz & D.J. Müller
Journal of Cell Science (2005) 118, 5315-5323.
Measuring cell adhesion forces of primary gastrulating cells from zebrafish using atomic force microscopy
P.-H. Puech, A. Taubenberger, F. Ulrich, M. Krieg, D.J. Müller & C.P. Heisenberg
Journal of Cell Science (2005) 118, 4199-4206.
