DeePhys: New machine-learning-assisted platform for electrophysiological phenotyping
Newly developed machine-learning-assisted platform DeePhys enables phenotypic electrophysiological screenings at the cellular and network level, identification of reliable features to classify healthy and mutant neuron lines, and comprehensive evaluation of pharmacological interventions.
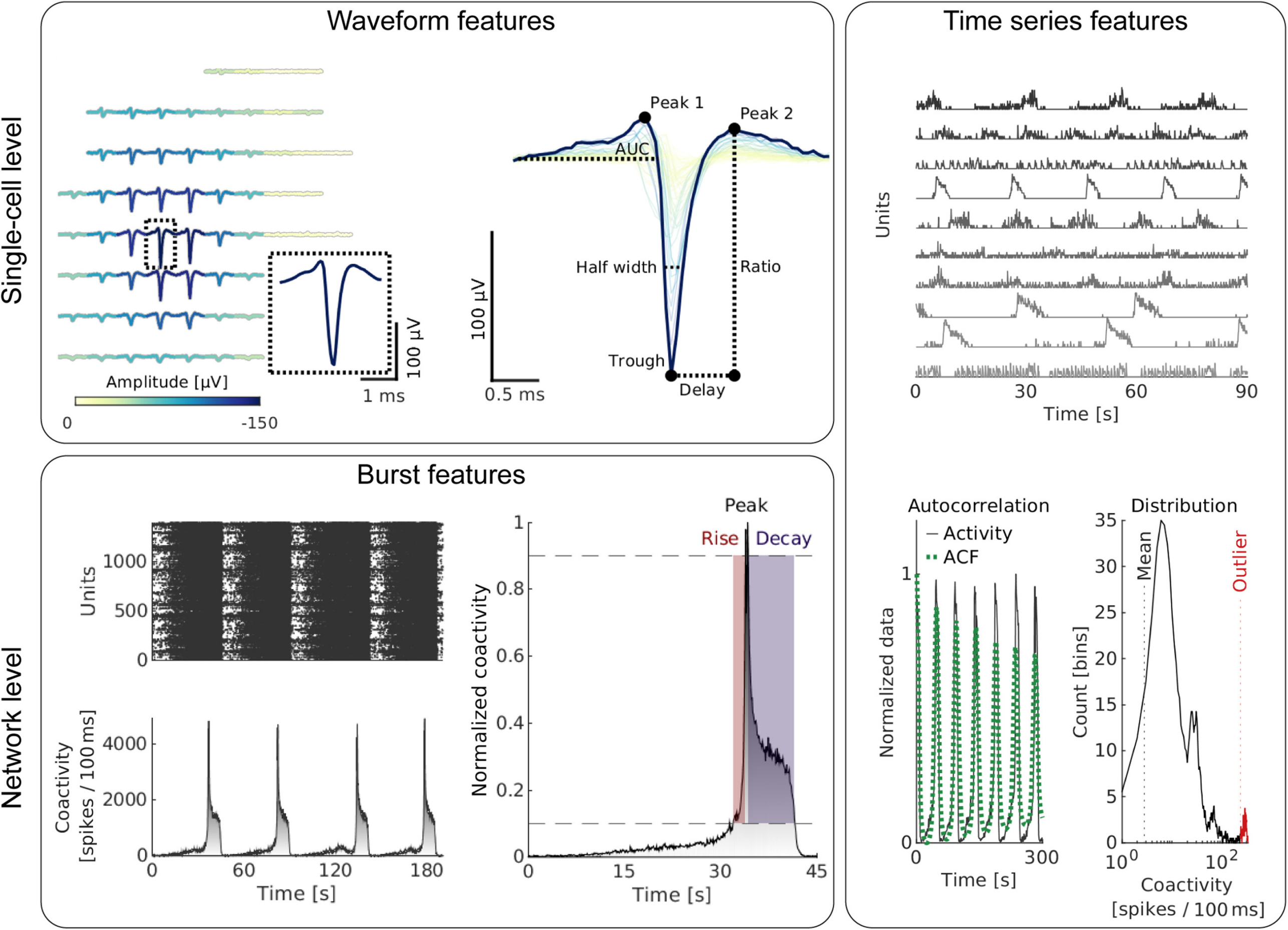

Reproducible functional assays to study in vitro neuronal networks represent an important cornerstone in the quest to develop physiologically relevant cellular models of human diseases. Therefore, we developed DeePhys, a MATLAB-based analysis tool for data-driven functional phenotyping of in vitro neuronal cultures, recorded by high-density microelectrode arrays (P. Hornauer et al., "DeePhys: A machine learning–assisted platform for electrophysiological phenotyping of human neuronal networks", Stem Cell Reports 2023 (DOI: 10.1016/j.stemcr.2023.12.008). DeePhys is a modular workflow that offers a range of techniques to extract features from spike-sorted data, allowing for the examination of functional phenotypes at the individual-cell and network levels, as well as across development. In addition, DeePhys incorporates the capability to integrate novel features and to use machine-learning-assisted approaches, which facilitates a comprehensive evaluation of pharmacological interventions. To illustrate its practical application, DeePhys was applied to human induced pluripotent stem cell–derived dopaminergic neurons obtained from both, patients and healthy individuals. We showcased how DeePhys enables phenotypic screenings.
