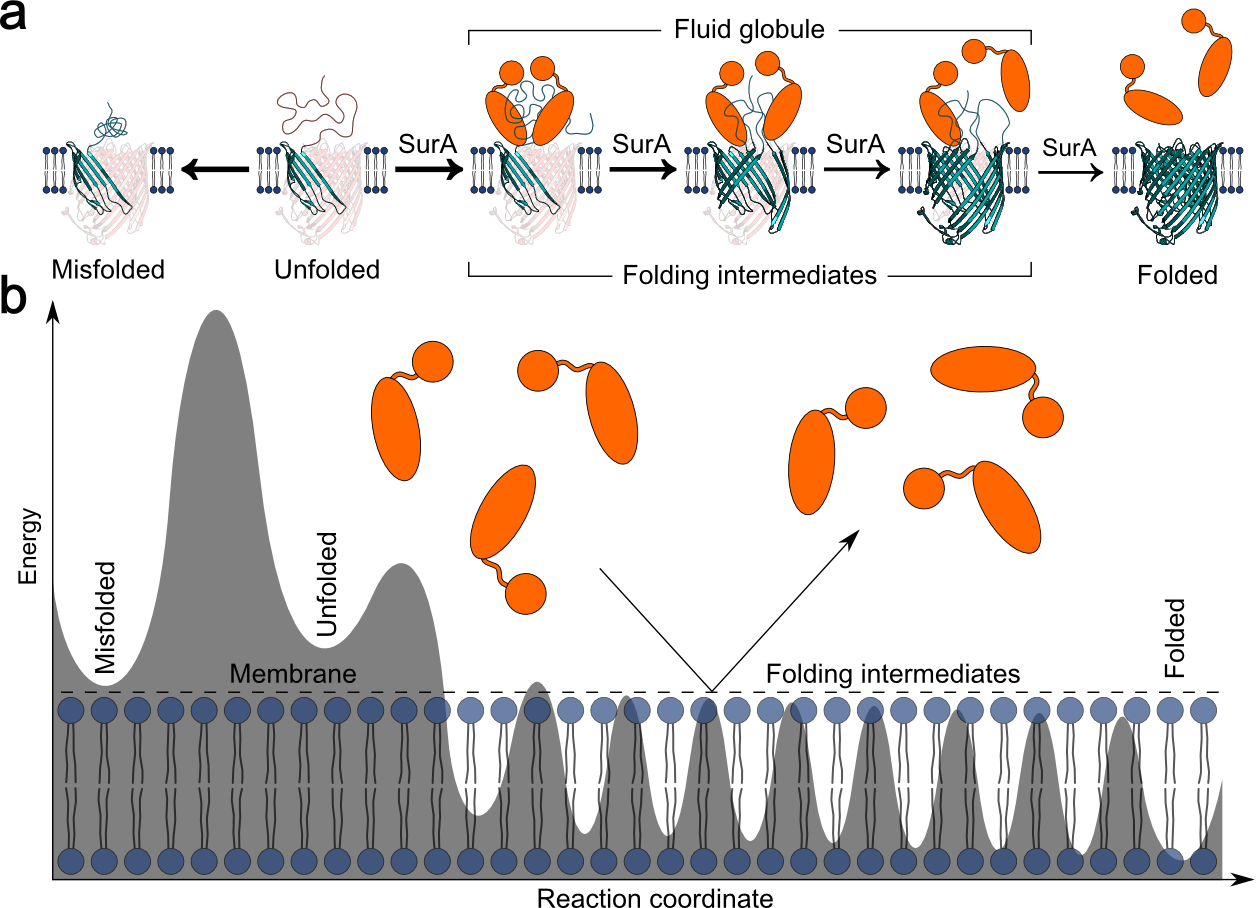
Observing the Folding of Single Proteins into Membranes
One of the great challenges for molecular and cell biologists is to understand how a polypeptide defines the three-dimensional structure of the a protein. It is even more difficult to understand this problem for membrane proteins because so little was known about what they looked like. In the recent years the situation has improved markedly as more membrane protein strutcures have been solved. However, it remains still largely unknown how a polypeptide itself or with the help of chaperones, translocons and insertases can insert and fold into the native membrane. We thus develop methods that allow to observe the unfolding and folding of single polypeptides into membranes at native-like conditions. Currently the resolution of these folding experiments allow us to observe the unfolding steps and folding steps of single secondary structures and how theses steps form the folding pathway towards the completely folded functional membrane protein.
Further reading
YidC assists the stepwise and stochastic folding of membrane proteins
T. Serdiuk, D. Balasubramaniam, J. Sugihara, S.A. Mari, H.R. Kaback & D.J. Muller
Nature Chemical Biology (2016) 11, 911-917.
Molecular plasticity of the human voltage-dependent anion channel embedded into a membrane
L. Ge, S. Villinger, S.A. Mari, K. Giller, C. Griesinger, S. Becker, D.J. Müller & M. Zweckstetter
Structure (2016) 24, 585-594.
Monitoring backbone hydrogen bond formation in β-barrel membrane protein folding
T. Raschle, P. Rios Flores, C. Opitz, D.J. Müller & S. Hiller
Angewandte Chemie International Edition (2016) 55, 5952-5955. external pageRead Morecall_madeexternal pagecall_made
Impact of holdase chaperones Skp and SurA on the folding of β-barrel outer membrane proteins
J. Thoma, B.M. Burmann, S. Hiller & D.J. Muller
Nature Structural and Molecular Biology (2015), 22, 795-802. external pageRead Morecall_madeexternal pagecall_made
Observing a lipid-dependent alteration in single lactose permeases
T. Serdiuk, J. Sugihara, S.A. Mari, H.R. Kaback & D.J. Müller
Structure (2015) 23, 754-761.
Action of the Hsp70 chaperone system observed with single proteins
J.M. Nunes, M. Mayer-Hartl, F.U. Hartl & D.J. Muller
Nature Communications (2015) 6, 6307.
Substrate-induced changes in the structural properties of LacY
T. Serdiuk, M.G. Madej, J. Sugihara, S. Kawamura, S.A. Mari, H.R. Kaback & D.J. Muller
Proc. Natl. Acad. Sci. USA (2014) 111, E1571-E1580.
The peptide transporter DtpA populates two alternate conformations from which inhibitor binding promotes one
C. Bippes, L. Ge, M. Meury, D. Harder, Z. Ucurum, H. Daniel, D. Fotiadis & D.J. Muller
Proc. Natl. Acad. Sci. USA (2013) 110, E3978-3986.
Mechanistic explanation of different unfolding behaviors observed for transmembrane and soluble β-barrel proteins
U. Hensen & D.J. Muller
Structure (2013) 21, 1317-1324.
Single-molecule force spectroscopy of G-protein coupled receptors
M. Zocher, Ch. Bippes, C. Zhang & D.J. Muller
Chemical Society Reviews (2013) 42, 7801-7815.
Structural, kinetic, and mechanical differences between dark-state rhodopsin and opsin
S. Kawamura, M. Gerstung, L. Colozo, J. Helenius, N. Beerenwinkel, P.S.H. Park & D.J. Muller
Structure (2013) 21, 426-437.
Cholesterol increases kinetic, energetic, and mechanical stability of the human β2 adrenergic receptor
M. Zocher, C. Zhang, G.F.S. Rassmussen, B.K. Kobilka & D.J. Muller
Proc. Natl. Acad. Sci. USA (2012) 109, E3463-3473.
Out but not in: β-strands shape the unfolding pathway but not the refolding of the large transmembrane β-barrel protein FhuA
J. Thoma, P. Bosshart, M. Pfreundschuh & D.J. Muller
Structure (2012) 20, 2185-2190.
Characterization of co-translationally formed nanodisc complexes with small multidrug transporters, proteorhodopsin and with the E. coli MraY translocase
C. Roos, M. Zocher, D.J. Muller, D. Munch, T. Schneider, H.G. Sahl, F. Scholz, J. Wachtveitl, Y. Ma, D. Proverbio, E. Henrich, V. Dötsch & F. Bernhard
Biochemical Biophysical Acta (2012) 1818, 3098-3106.
Ligand-specific interactions modulate kinetic, energetic and mechanical properties of the human β2 adrenergic receptor
M. Zocher, J.J. Fung, B.K. Kobilka & D.J. Muller
Structure (2012) 20, 1391-1402.
Structural, energetic and mechanical perturbations in a rhodopsin mutant that causes congenital stationary night blindness
S. Kawamura, A.L. Colozo, L. Ge, D.J. Muller & P.S.H. Park
Journal of Biological Chemistry (2012) 287, 21826-21835.
KpOmpA a transmembrane protein anchoring the outer membrane of Klebsiella pneumoniae unfolds and refolds in response to tensile load
P. Bosshart, I. Iordanov, C. Garzon-Coral, P. Demange, A. Engel, A. Milon & D.J. Muller
Structure (2012) 20, 121-127.
Competing interactions stabilize pro- and anti-aggregant conformations of human Tau
S. Wegmann, J. Schöler, C.A. Bippes, E. Mandelkov & D.J. Muller
Journal of Biological Chemistry (2011) 286, 20512-20524.
One β-hairpin after the other: Exploring refolding pathways and kinetics of the transmembrane β-barrel protein OmpG
M. Damaghi, S. Koester, C.A. Bippes, Ö. Yildiz & D.J. Muller
Angewandte Chemie International Edition (2011) 50, 7422-7424.
High-resolution atomic force microscopy and spectroscopy of native membrane proteins
Ch. Bippes & D.J. Muller
Reports on Progress in Physics (2011) 74, 086601.
Conservation of molecular interactions stabilizing bovine and mouse rhodopsin
S. Kawamura, A.T. Colozo, D.J. Müller & P.S.H. Park
Biochemistry (2010) 49, 10412-10420.
A ”force buffer” protecting immunoglobulin titin
J.A. Nunes, U. Hensen, L. Ge, M. Lipinsky, J. Helenius, H. Grubmüller & D.J. Müller
Angewandte Chemie International Edition (2010) 49, 3528-3531.
Dual energy landscape: The functional state of the outer-membrane β-barrel protein OmpG molds its unfolding energy landscape
M. Damaghi, K.T. Sapra, S. Köster, Ö. Yildiz, W. Kühlbrandt & D.J. Müller
Proteomics (2010) 10, 4151-4162.
Quantifying interactions that switch the β-barrel membrane protein OmpG from the open to the closed state
M. Damaghi, C.A. Bippes, S. Köster, S.A. Mari, Ö. Yildiz, W. Kühlbrandt & D.J. Müller
Journal of Molecular Biology (2010) 397, 878-882.
Probing the interactions of carboxy-atractyloside and atractyloside with the yeast mitochondrial ADP/ATP carrier
A. Kedrov, A.M. Hellawell, A. Klosin, R.B. Broadhurst, E.R.S. Kunji & D.J. Müller
Structure (2010) 18, 39-46.
One β-hairpin after the other: Exploring mechanical unfolding pathways of the transmembrane β-barrel protein OmpG
K.T. Sapra, M. Damaghi, S. Köster, Ö. Yildiz, W. Kühlbrandt & D.J. Müller
Angewandte Chemie International Edition (2009) 48, 8306-8308.
Modulation of molecular interactions and function by rhodopsin palmitylation
P.S.H. Park, K.T. Sapra, B. Jastrzebska, T. Maeda, A. Maeda, W. Pulawski, M. Kono, J. Lem, R.K. Crouch, S. Filipek, D.J. Müller & K. Palczewski
Biochemistry (2009) 48, 4294-4304.
Substrate binding tunes conformational flexibility and kinetic stability of an amino acid transporter
C.A. Bippes, A. Zeltina, F. Casagrande, M. Ratera, M. Palacin, D.J. Müller & D. Fotiadis
Journal of Biological Chemistry (2009) 284, 18651-18663.
AFM: A nanotool in membrane biology
D.J. Müller
Biochemistry (2008) 47, 7986-7998.
High-throughput single-molecule force spectroscopy for membrane proteins
P.D Bosshart, F. Casagrande, P.L.T.M. Frederix, M. Ratera, C.A Bippes, D.J. Müller, M.l Palacin, A. Engel & D. Fotiadis
Nanotechnology (2008) 19, 384014.
Fully automated single-molecule force spectroscopy for screening applications
J. Struckmeier, R. Wahl, M. Leuschner, J.M. Nunes, H. Janovjak, U. Geisler, G. Hofmann, T. Jähnke &D.J. Müller
Nanotechnology (2008) 19, 384020.
Transducer binding establishes localized interactions that tune sensory rhodopsin II
D. Cisneros, L. Oberbarnscheidt, A. Pannier, J.P. Klare, J. Helenius, M. Engelhard, F. Oesterhelt & D.J. Müller
Structure (2008) 16, 1206-1213.
Role of extracellular glutamic acids in the stability and energy landscape of bacteriorhodopsin
K.T. Sapra, J. Doehner, V. Renugolpalkrishan, E. Padros & D.J. Müller
Biophysical Journal (2008) 95, 3407-3418.
From valleys to ridges: Exploring the energy landscape of single membrane proteins
H. Janovjak, K.T. Sapra, A. Kedrov & D.J. Müller
ChemPhysChem (2008) 9, 954-956.
Folding and assembly of proteorhodopsin
A.L. Klyszejko, S. Shastri, S.A. Mari, H. Grubmüller, D.J. Müller & C. Glaubitz
Journal of Molecular Biology (2008) 376, 35-41.
Mechanical properties of bovine rhodopsin and bacteriorhodopsin dictate energy landscape and guide conformational changes
K.T. Sapra, P.S.-H. Park, K. Palczewski & D.J. Müller
Langmuir (2008) 24, 1330-1337.
Point mutations of membrane protein alter energy landscape and unfolding pathways
K.T. Sapra, P. Balasubramanian, D. Labudde, J. Bowie & D.J. Müller
Journal of Molecular Biology (2008) 376, 1076-1090.
Examining the dynamic energy landscape of an antiporter upon inhibitor binding
A. Kedrov, M. Appel, H. Baumann, Ch. Ziegler & D.J. Müller
Journal of Molecular Biology (2008) 375, 1258-1266.
Atomic force microscopy and spectroscopy of native membrane proteins
D.J. Müller & A. Engel
Nature Protocols (2007) 2, 2191-2197.
Free energy of membrane protein unfolding derived from single-molecule force measurements
J. Preiner, H. Janovjak, C. Rankl, H. Knaus, D.A. Cisneros, A. Kedrov, F. Kienberger, D.J. Müller & P. Hinterdorfer
Biophysical Journal (2007) 93, 930-937.
Stabilizing effect of Zn2+ in native bovine rhodopsin
P.S.-H. Park, K.T. Sapra, M. Koliński, S. Filipek, K. Palczewski & D.J. Müller
Journal of Biological Chemistry (2007) 282, 11377-11385.
Transmembrane helices have rough energy surfaces
H. Janovjak, H. Knaus & D.J. Müller
Journal of the American Chemical Society (2007) 129, 246-247.
Detecting molecular interactions that stabilize, activate and guide ligand-binding of the sodium/proton antiporter MjNhaP1 from Methanococcus jannaschii
A. Kedrov, S. Wegmann, S. Smiths, P. Goswami, H. Baumann & D.J. Müller
Journal of Structural Biology (2007) 159, 290-301.
Deciphering molecular interactions of native membrane proteins by single-molecule force spectroscopy
A. Kedrov, H. Janovjak, K.T. Sapra & D.J. Müller
Annual Review of Biophysics and Biomolecular Structure (2007) 36, 233-260.
Differentiating ligand and inhibitor interactions of a single antiporter
A. Kedrov, Ch. Ziegler & D.J. Müller
Journal of Molecular Biology (2006) 362, 925-932.
Bacteriorhodopsin folds into the membrane against an external force
M. Kessler, K. Gottschalk, H. Janovjak, D.J. Müller & H.E. Gaub
Journal of Molecular Biology (2006) 357, 644-654.
Characterizing molecular interactions in different bacteriorhodopsin assemblies by single-molecule force spectroscopy
T. Sapra, H. Besir, D. Oesterhelt & D.J. Müller
Journal of Molecular Biology (2006) 355, 640-650.
Direct measurement of single-molecule visco-elasticity in AFM force-extension measurements
Ch. Bippes, A. Humphris, D.J. Müller & H. Janovjak
European Biophysical Journal (2006) 35, 287-292.
Observing folding pathways and kinetics of a single membrane protein
A. Kedrov, H. Janovjak, Ch. Ziegler, W. Kühlbrandt & D.J. Müller
Journal of Molecular Biology (2006) 355, 2-8.
Locating ligand binding and activation of a single antiporter
A. Kedrov, M. Krieg, Ch. Ziegler, W. Kühlbrandt & D.J. Müller
Embo reports (2005) 6, 668-674.
Complex stability of single proteins explored by forced unfolding experiments
H. Janovjak, T.K. Sapra & D.J. Müller
Biophysical Journal (2005) 88, L37-L39.
Probing different origins of potential barriers stabilizing the membrane proteins halorhodopsin and bacteriorhodopsin
D. Cisneros, D. Oesterhelt & D.J. Müller
Structure (2005) 13, 235-242.
Dynamic response of single bacteriorhodopsins, determined by single-molecule force modulation spectroscopy
H. Janovjak, D.J. Müller & A. Humphris
Biophysical Journal (2005) 88, 1423-1431.
Probing the energy landscape of the membrane protein bacteriorhodopsin
H. Janovjak, J. Struckmeier, M. Hubain, A. Kedrov, M. Kessler & D.J. Müller
Structure (2004) 12, 871-879.
Controlled unfolding and refolding of a single sodium-proton antiporter
A. Kedrov, Ch. Ziegler, H. Janovjak, W. Kühlbrandt & D.J. Müller
Journal of Molecular Biology (2004) 340, 1143-1152.
Temperature dependent unfolding of bacteriorhodopsin
H. Janovjak, M. Kessler, D. Oesterhelt, H. Gaub & D.J. Müller
EMBO Journal (2003) 22, 5220-5229.
Determining molecular forces that stabilize human aquaporin-1
C. Moeller, D. Fotiadis, K. Suda, A. Engel, M. Kessler & D.J. Müller
Journal of Structural Biology (2003) 142, 369-378.
Stability of bacteriorhodopsin α-helices and loops analyzed by single molecule force spectroscopy
D.J. Müller, M. Kessler, F. Oesterhelt, C. Moeller, D. Oesterhelt & H. Gaub
Biophysical Journal (2002) 83, 3578-3588.
Conformations of the rhodopsin third cytoplasmic loop grafted onto bacteriorhodopsin
J.B. Heymann, M. Pfeiffer, V. Hildebrandt, D. Fotiadis, B. de Groot, R. Kabak, A. Engel, D. Oesterhelt & D.J. Müller
Structure (2000) 8, 643-653.
Unfolding pathways of individual bacteriorhodopsins
F. Oesterhelt, D. Oesterhelt, M. Pfeiffer, A. Engel, H. Gaub & D.J. Müller
Science (2000) 288, 143-146. [Commentary by J.G. Forbes & G.H. Lorimer ‘Unravelling a membrane protein’ Science (2000) 288, 63-64]
