Functional Imaging of Membrane Receptors and Cellular Membranes
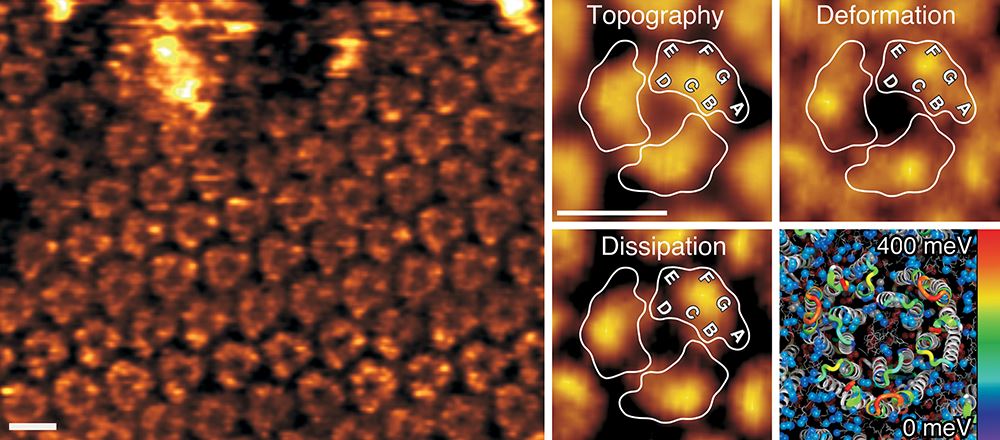
A current challenge in the life sciences is to understand how biological systems change their structural, biophysical and chemical properties to adjust functionality. Addressing this issue has been severely hampered by the lack of methods capable of imaging biosystems at high resolution while simultaneously mapping their multiple properties. Recent developments in force-distance curve–based atomic force microscopy (FD-based AFM) now enable researchers to combine (sub)molecular imaging with quantitative mapping of physical, chemical and biological interactions. We thus develop and apply advanced FD-based AFM tools for the quantitative multiparametric characterization of complex cellular and biomolecular systems under physiological conditions. In this focus of our research we largely apply the multiparametric high-resolution AFM to characterize membrane proteins, cell membrane patches and living cells.
Publications on the topic
Nanomechanical mapping of first binding steps of a virus to animal cells
D. Alsteens, R. Newton, R. Schubert, D. Martinez-Martin, M. Delguste, B. Roska & D.J. Muller
Nature Nanotechnology (2016) in press.
SAS-6 engineering reveals interdependency between cartwheel and microtubule wall in determining centriole architecture
M. Hilbert, A. Noga, D. Frey, V. Hamel, P. Guichard, S.H.W. Kraatz, M. Pfreundschuh, S. Hosner, I. Flückiger, R. Jaussi, M.M. Wieser, K.M. Thieltges, X. Deupi, D.J. Müller, R.A. Kammerer, P. Gönczy, M. Hirono & M.O. Steinmetz
Nature Cell Biology (2016) 18, 393-403.
Quantifying and identifying two ligand-binding sites while imaging native human membrane receptors by AFM
M. Pfreundschuh, D. Alsteens, R. Wieneke, C. Zhang, S.R. Coughlin, R. Tampé, B.K. Kobilka & D.J. Müller
Nature Communications (2015) 6, 8857. external pageonlinecall_made
Directly observing the lipid-dependent self-assembly and pore-forming mechanism of the cytolytic toxin listeriolysin O
E. Mulvihill, K. van Pee, S.A. Mari, D.J. Müller & Ö. Yildiz
Nano Letters (2015) 15, 6965-6973 external pageonlinecall_made
Imaging G protein-coupled receptors while quantifying their ligand-binding free-energy landscape
D. Alsteens, M. Pfreundschuh, C. Zhang, P. Spoerri, S.R. Coughlin, B.K. Kobilka & D.J. Müller
Nature Methods (2015) 12, 845-851.
Localizing chemical groups while imaging single native proteins by high-resolution AFM
M. Pfreundschuh, D. Alsteens, M. Hilbert, M.O. Steinmetz & D.J. Müller
Nano Letters (2014) 14, 2957-2964.
Multiparametric high-resolution imaging of native proteins by force-distance curve–based AFM
M. Pfreundschuh, D. Martinez-Martin, E. Mulvihill, S. Wegmann & D.J. Muller
Nature Protocols (2014) 9, 1113-1130.
Quantitative imaging of the electrostatic field and potential generated by a transmembrane protein at sub-nanometer resolution
M. Pfreundschuh, U. Hensen & D.J. Muller
Nano Letters (2013) 13, 5585-5593.
Multi-parametric imaging of biological systems by force-distance curve-based AFM
Y.F. Dufrene, D. Martínez-Martín, I. Medalsy, D. Alsteens & D.J. Muller
Nature Methods (2013) 10, 847-854.
Nanomechanical properties of proteins and membranes depend on loading rate and electrostatic interactions
I. Medalsy & D.J. Muller
ACS NANO (2013) 7, 2642-2650.
The fuzzy coat of pathological human Tau fibrils is a polyelectrolyte polymer brush
S. Wegmann, I. Medalsy, E. Mandelkow & D.J. Muller
Proc. Natl. Acad. Sci. USA (2013) 110, E313-321.
Quantifying chemical and physical properties of native membrane proteins at molecular resolution by force-volume AFM
I. Medalsy, U. Hensen & D.J. Muller
Angewandte Chemie International Edition (2011) 50,12103-12108.
Force probing surfaces of living cells to molecular resolution
D.J. Müller, J. Helenius, D. Alsteens & Y.F. Dufrene
Nature Chemical Biology (2009) 5, 383-390.
Atomic force microscopy as a multifunctional molecular toolbox in nanobiotechnology
D.J. Müller & Y. Dufrene
Nature Nanotechnology (2008) 3, 261-269.
Atomic force microscopy and spectroscopy of native membrane proteins
D.J. Müller & A. Engel
Nature Protocols (2007) 2, 2191-2197.
Imaging the electrostatic potential of transmembrane channels: Atomic probe microscopy of OmpF porin
A. Philippsen, W. Im, A. Engel, T. Schirmer, B. Roux & D.J. Müller
Biophysical Journal (2002) 82, 1667-1676.
